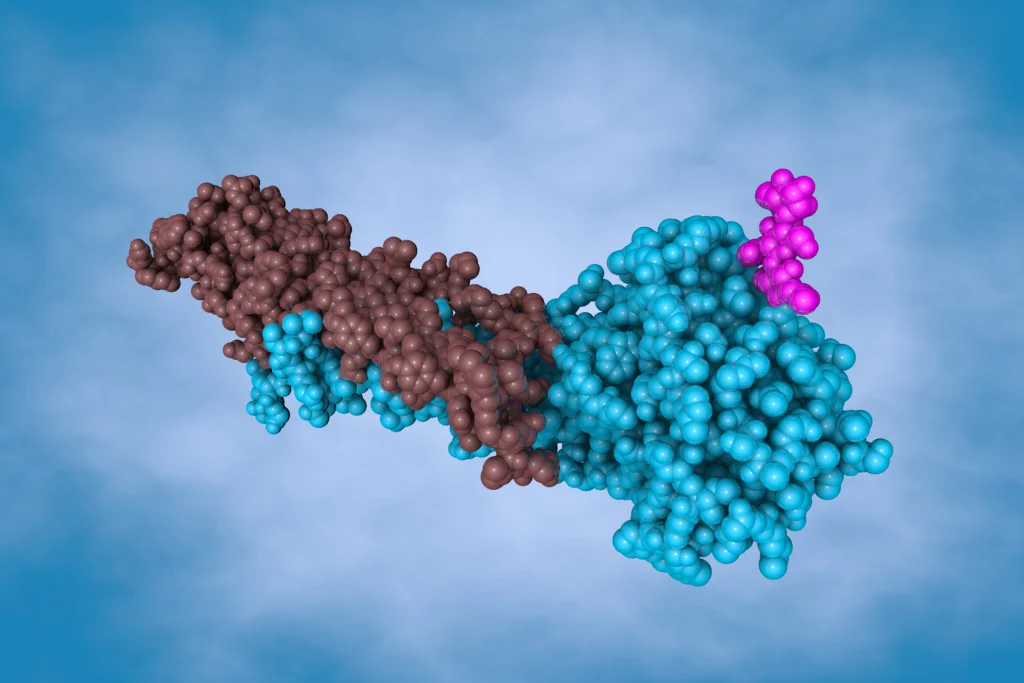
What Are Molecular Glues
Molecular glues represent a transformative modality in drug discovery, enabling the modulation of protein function through induced proximity. Unlike traditional inhibitors, molecular glues facilitate or stabilize protein–protein interactions (PPIs), often leading to novel biological outcomes such as complex stabilization, altered signaling, or inhibition of pathological interactions—without necessarily inducing protein degradation (1, 2, 3).
Recent advances have brought non-degradative molecular glues into clinical development, expanding their therapeutic potential beyond targeted protein degradation. Notable examples include:
– RMC-6291 (4) by Revolution Medicines: A KRasG12C-targeting molecular glue that stabilizes an inactive ternary complex between KRas and cyclophilin A (CypA), bypassing the need for a druggable pocket on KRas. This approach enables inhibition of oncogenic signaling in KRas-driven cancers such as lung and colorectal cancer.
– NST-628 (5) by Nested Therapeutics: A brain-penetrant molecular glue that stabilizes RAF-MEK complexes in the RAS-MAPK pathway. It is currently in Phase 1 trials for solid tumors with RAS-MAPK mutations, including melanoma and pancreatic cancer. NST-628 (5) exemplifies a non-degradative glue that inhibits pathway reactivation by locking signaling complexes in inactive conformations.
These examples highlight the growing clinical relevance of non-degradative molecular glues and underscore their potential to address previously undruggable targets with precision and versatility.
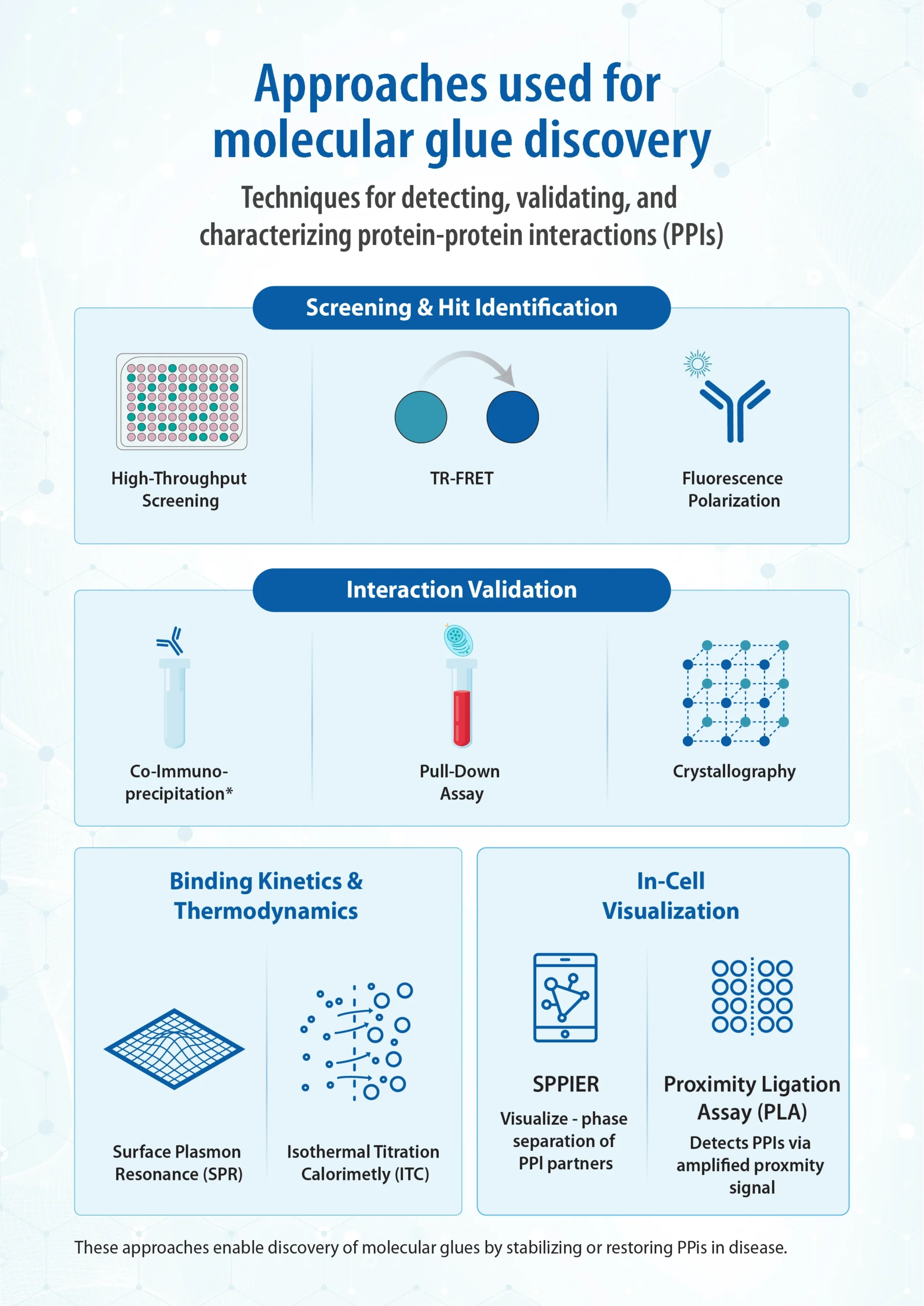
Approaches used for Molecular Glue Discovery
The discovery and characterization of molecular glues rely on a diverse array of biological assays designed to detect, validate, and understand protein–protein interactions (PPIs). These tools are essential for identifying compounds that modulate PPIs either by stabilizing or restoring them, often in disease-relevant contexts. High-throughput screening techniques such as TR-FRET and fluorescence polarization are commonly used to identify initial hits, while co-immunoprecipitation and pull-down assays confirm direct interactions (1, 2, 3). Structural methods like crystallography and cryo-electron microscopy (Cryo-EM) provide atomic-level insights into binding interfaces and glue-induced conformational changes. To further characterize binding kinetics and thermodynamics, surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) are employed. For in-cell visualization of PPIs, advanced techniques such as Separation of Phases-based Protein Interaction Reporter (SPPIER) and proximity ligation assays (PLA) offer valuable spatial and temporal resolution. The following table summarizes recent examples of molecular glue discovery using pre-selected presenter and target proteins, highlighting the various strategies and assay platforms employed to identify and characterize these unique modulators of protein–protein interactions. Wherever feasible, ITC is used to quantify binding and cooperativity while keeping the workflow light.
Recent Examples of Molecular Glue Discovery
Non-degradative molecular glues illustrate multiple use cases: stabilizing inactive signaling complexes to blunt pathway reactivation; restoring weakened tumor suppressor interactions (e.g., SMAD4–SMAD3) that are compromised by disease variants; and altering oligomeric states in viral proteins to reduce replication competence. Screening typically starts with rapid, mix-and-read assays (TR-FRET/Alpha or fluorescence polarization) to identify interface stabilizers, moves to biophysics (including isothermal titration calorimetry) to quantify thermodynamics and cooperativity, and then to cellular confirmation (e.g., co-immunoprecipitation or NanoBRET) to check whether induced proximity holds in context. Functional assays and pathway markers then link ternary complex formation to phenotypic outcomes. This progression keeps candidates honest and decision-ready.
Target PPI | Assay Type | Key Findings | Reference |
SMAD4/SMAD3 | TR-FRET | Identified Ro-31-8220 and Go-6983 as glues restoring mutant SMAD4–SMAD3 interaction | Tang et al., 2021 |
BCL6/BCOR | Fluorescence Polarization | Discovered glues that inhibit BCL6 and promote its degradation | Kerres et al., 2017 |
PCNA Trimers | Structure-Based Docking | Found compounds that stabilize PCNA trimers and inhibit monomer dissociation | Dillehay KL, et al. 2015 |
MERS-CoV Nucleocapsid Protein | Crystallography + Docking | Identified glue that induces oligomerization and inhibits viral replication | Xu et al., 2025 |
RAF–MEK Complexes | AlphaLISA, HTRF, SPR | NST-628 stabilizes RAF–MEK, inhibits MEK phosphorylation, effective in RAS tumors | Ryan et al., 2024 |
14-3-3σ with CRAF, TAZ, ERα | NanoBRET | Characterized glues stabilizing 14-3-3σ interactions in live cells | Vickery et al., 2024 |
These examples illustrate the diverse and innovative strategies employed in molecular glue discovery, emphasizing the critical role of working with teams that possess deep scientific expertise and broad technological capabilities. Syngene is well positioned to support such efforts, offering a comprehensive suite of assay platforms—including TR-FRET, fluorescence polarization, structure-based docking, AlphaLISA, SPR, HTRF, and NanoBRET—that enable the identification, validation, and optimization of molecular glues across a wide range of target classes and therapeutic areas. These assay platforms are complemented by Syngene’s strong capabilities in computational chemistry, structural biology, structure-based drug design (SBDD), and virtual screening which are increasingly vital in the rational discovery and optimization of molecular glues. These in silico strategies, when integrated with assay biology, significantly enhance the efficiency and success rate of molecular glue discovery campaigns.
Advantages of an Integrated Drug Discovery Approach
An integrated drug discovery approach, especially when guided by experts with decades of experience, offers a powerful and holistic framework for advancing therapeutic candidates. It enables seamless collaboration across cross-functional teams, ensuring continuity from target validation through to candidate nomination. Coordinated workflows and real-time data sharing accelerate timelines and improve decision-making, while the synergy between biology, chemistry, DMPK and pharmacology fosters enhanced innovation and creative problem-solving. Integrated data analysis allows for early identification of potential liabilities, significantly mitigating risk. Moreover, this model provides strategic flexibility, allowing teams to pivot based on emerging data and evolving project needs. Such an approach is particularly advantageous for complex modalities like molecular glues, where iterative design and deep mechanistic understanding are essential for success.
Conclusion
Molecular glues represent a versatile and powerful modality in modern drug discovery, extending the reach of small molecules into complex protein networks by harnessing induced proximity. Success depends on clear, orthogonal evidence: rapid screening to find signal, biophysics to anchor thermodynamics and cooperativity, structural snapshots to rationalize binding, and cellular confirmation to ensure biological relevance. As case studies continue to grow, disciplined, mechanism-led workflows will help in prioritizing, optimizing, and translating molecular glues into impactful therapies. We recommend routine isothermal titration calorimetry checks alongside fluorescence polarization and co-immunoprecipitation to align mechanism with potency.