At a glance
A global biopharma sponsor developing a novel bispecific nanobody needed a fit-for-purpose PK assay that could accurately measure drug concentrations across a wide exposure range. The program depended on reliable bioanalysis to quantify very low terminal-phase levels as well as high peak concentrations after dosing. Syngene’s large molecule bioanalysis team redesigned the calibration curve to achieve a dynamic range of 1.00–1000 ng/mL and resolved issues with assay drift and systemic negative bias. The optimized method showed a repeat rate of ~1%, enabling robust pharmacokinetic profiling and confident dose selection under GLP compliance frameworks.
Background: large molecule bioanalysis for a bispecific nanobody
The sponsor’s investigational drug was a bispecific nanobody, a compact antibody-derived construct engineered to bind two different targets at the same time. These molecules combine high specificity with faster systemic clearance compared with conventional monoclonal antibodies. This profile offers advantages for targeted therapy but places extra demands on bioanalysis, especially for clinical PK characterization.
For this program, the sponsor needed a PK assay that could support early and later clinical studies. The assay had to quantify nanobody concentrations at very low levels to characterize the terminal phase, handle high exposure levels without losing accuracy or requiring frequent dilutions, and maintain consistent performance across runs to avoid repeat testing and data delays. A single, well-optimized bioanalysis method was preferred over multiple assays to simplify clinical operations and reduce variability.
Challenge in ligand binding assay development and PK assay performance
During early ligand binding assay development, the transferred method showed several performance issues that limited its suitability for clinical bioanalysis. The dynamic range was not adequate for low-concentration measurements, and the assay could not reliably quantify the lowest expected concentrations, which risked underestimating exposure in the terminal phase.
There was also noticeable drift across the plate, with signal changes over time and position affecting sensitivity and leading to inconsistent recovery of quality-control (QC) samples. In addition, regression analysis showed a systemic negative bias in spike-recovery results, with QC samples tending to read lower than their nominal values. This raised concerns about the underestimation of true drug levels.
Taken together, these problems increased the risk of repeat analysis and could compromise the reliability of PK data for clinical decision-making. For the sponsor, this meant potential delays in interpreting concentration–time profiles and more uncertainty during dose selection, even though overall bioanalysis capacity was available.
Syngene’s solution: stabilizing bioanalysis and extending PK assay range
Syngene’s large molecule bioanalysis team adopted a stepwise experimental strategy to stabilize the method and extend the dynamic range so it would function as a fit-for-purpose PK assay for the bispecific nanobody.
Redesigning the standard curve
The first step was to redesign the standard curve to extend the quantifiable range. Calibration levels and signal response were optimized so that the dynamic range covered 1.00–1000 ng/mL. This change allowed accurate measurement of both high and low nanobody concentrations within a single run. As a result, the need for additional dilutions was reduced and overall re-analysis was minimized, improving overall bioanalysis efficiency.
Addressing drift and bias in ligand binding assay development
The team then focused on resolving plate drift and negative bias as part of systematic ligand binding assay development. Signal trends, instrument response, and spike-recovery data were reviewed to understand contributions from reagents and plate-handling patterns. Based on these insights, capture and detection reagents were titrated to provide a more stable signal across the entire plate, and mixing steps were standardized with revised well-loading sequences to minimize temporal drift and edge effects.
With these changes, spike-recovery bias improved from an initial range of approximately –15% to –65% to a narrower, acceptable band around –4% to –8%. This level of performance was consistent with a fit-for-purpose PK assay and reduced the likelihood of systematic underestimation.
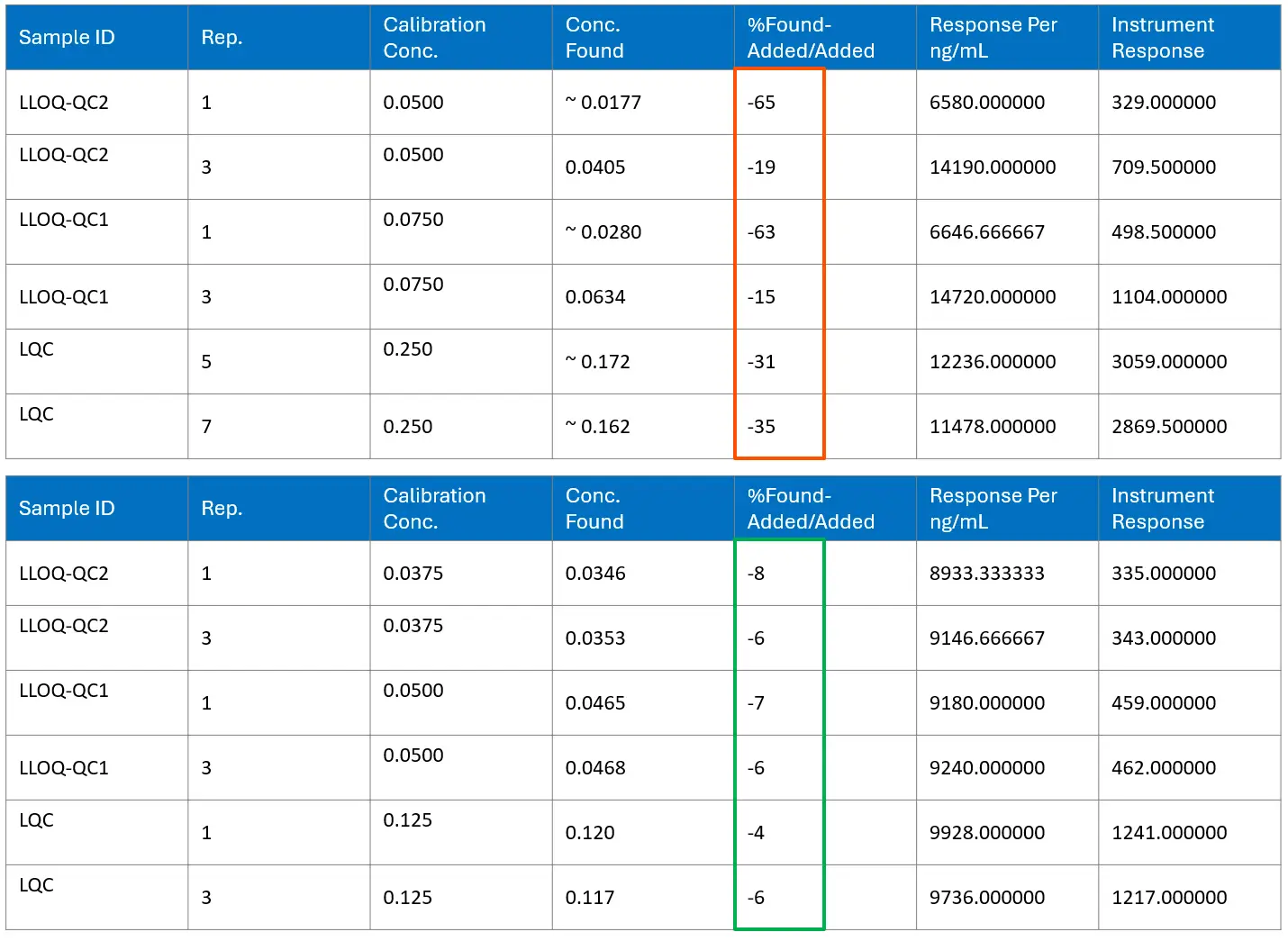
Clinical application, GLP compliance, and PK profiling
After optimization, the method was validated and applied to clinical samples as part of routine large molecule bioanalysis. The 1.00–1000 ng/mL dynamic range enabled accurate measurement of high exposure samples without compromising sensitivity at the low end. Drug levels as low as 1.43 ng/mL were reliably detected, confirming that the assay could capture late time points and end-of-trial concentrations.
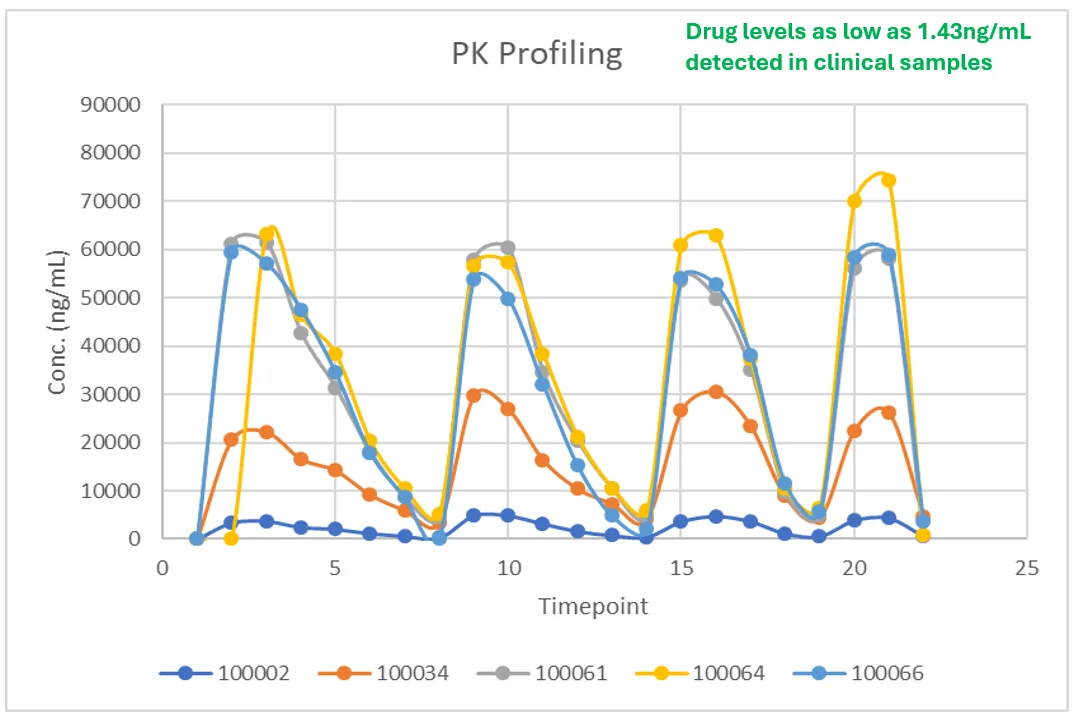
Across the clinical study, the repeat rate was approximately 1%, reflecting a robust method with limited need for re-analysis. This low repeat rate helped conserve samples, shortened reporting timelines, and improved the overall quality of bioanalysis data available to the project team. The resulting concentration–time profiles across subjects were smooth and internally consistent, enabling estimation of parameters such as Cmax and AUC with confidence. This supported clinical and regulatory discussions on exposure and informed dose selection for subsequent study phases, all under GLP compliance expectations for regulated bioanalysis.
Outcome and sponsor value
The optimized bioanalysis method delivered clear benefits for the sponsor’s bispecific nanobody program. It extended the usable assay range from 1.00 to 1000 ng/mL, covering both peak and terminal concentrations within a single run. Accuracy improved, with spike-recovery results mostly between –4% and –8% of nominal values, which reduced systemic negative bias. The method reliably detected drug levels down to 1.43 ng/mL, supporting detailed characterization of the elimination phase.
The overall repeat rate of around 1% reduced re-analysis, saved time, and limited additional sample use. By resolving assay drift and negative bias and unifying development, validation, and clinical sample analysis within one large molecule bioanalysis team, Syngene generated robust PK data that supported dose selection and future clinical planning. The sponsor received a fit-for-purpose PK assay that aligned with regulatory expectations and could be extended into later phases with minimal rework.







