Rise of the CHO cell line
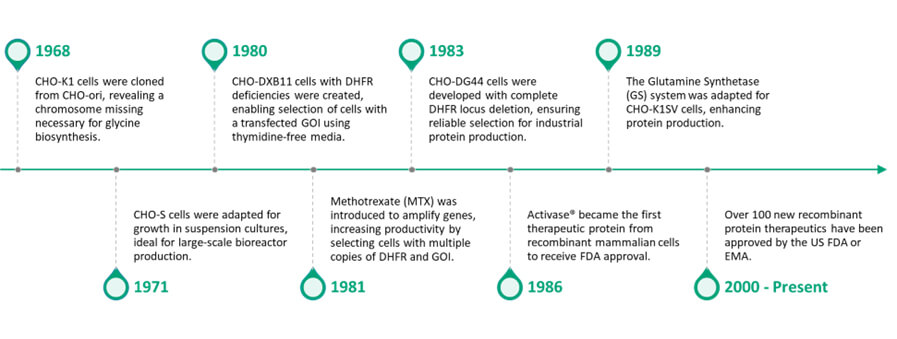
Chinese Hamster Ovary (CHO) cells have played a pivotal role in the production of biopharmaceutical proteins since their introduction in the late 20th century. Initially derived from the ovaries of Chinese hamsters in the 1950s, CHO cells were first utilized for recombinant protein production in the 1980s. Their significance in biopharmaceutical manufacturing stems from their ability to grow in suspension cultures, high adaptability to various growth conditions, and capacity for post-translational modifications that are critical for the functionality of therapeutic proteins. CHO cells are particularly valued for their ability to produce complex glycoproteins, which are essential for the efficacy and safety of many biologics, including monoclonal antibodies and therapeutic proteins.
Overtime, CHO cells have become the gold standard in cell line development, contributing to the successful commercialization of numerous life-saving treatments. Their reliability, scalability, and established regulatory pathways further underscore their importance in the biopharmaceutical sector, making them an integral component in the quest to meet global health challenges.
Figure 1: How CHO cells evolved overtime to become the gold standard in cell line development
Bottlenecks in traditional cell line development
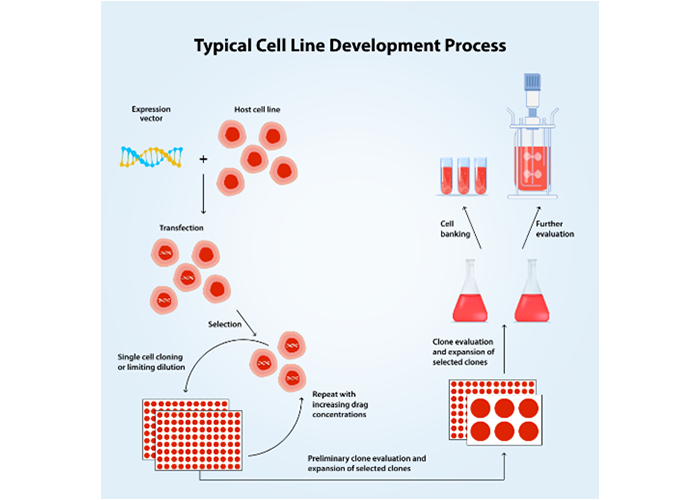
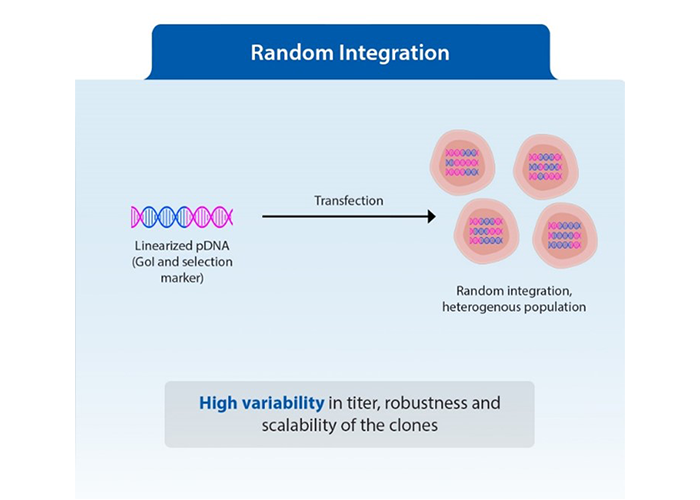
The traditional cell line development (CLD) workflow generally starts with the transfection of the gene of interest into the chosen cell line, followed by selection, limiting dilution, and cell banking. A major bottleneck in this traditional CLD workflow is random integration. Researchers have little control over where the gene integrates, necessitating extensive screening of numerous clones to identify the best producers. Additionally, assessing quality parameters across all clones is often impractical.
Consequently, finding the ideal clone with high titers and the right quality parameters can prove to be a complex and time-intensive endeavor.
The traditional approach has two major shortcomings– low integration efficiency and heterogeneity in gene expression due to position effects. This heterogeneity arises when the gene integrates at different sites in the genome, leading to varied expression levels among clones. To mitigate these position effects, several technologies have emerged.
Figure 2: Typical cell line development process
Approaches to mitigate the position effect
One of the approaches is the use of insertion of genetic elements into the vector backbone which have shown to reduce position effects and enhance stability by preventing transgene silencing and ensuring consistent, stable, high-level gene expression, regardless of the chromosomal integration site, another approach is, platform expression systems employing targeted integration can further reduce the impact of position effects. These systems specify a “landing pad” in the genome for gene insertion, minimizing variability in gene expression by controlling integration sites.
Traditional homologous recombination has been used for specific gene integration, while newer methods, such as nuclease-mediated targeting, utilize nucleases to create double-strand breaks, facilitating precise gene insertion. Recombinases—proteins like Cre, PhiC31, FLP, and BxB1—enable accurate integration at sequence-specific sites by catalyzing recombination between predefined locations. CRISPR-Cas technology has further enhanced gene editing capabilities. It allows for rapid and efficient modifications that enable precise insertion at single or multiple loci in mammalian genomes, including CHO cells. The use of multi-copy vectors has also been shown to increase the number of integrated gene copies, contributing to higher overall yields.
Harnessing the power of transposons
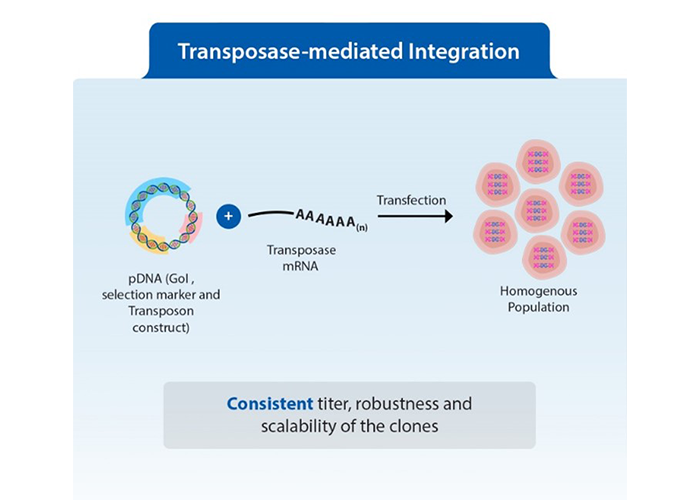
Transposon-mediated semi-targeted integration represents a significant advancement in the cell line development (CLD) workflow, particularly in addressing the inefficiency of traditional methods. By utilizing a transposase system, this approach allows for the semi-targeted integration of transgenes into the genome, dramatically reducing the number of clones that need to be screened.
Figure3: Random integration versus transposase-mediated integration of transgene into genomes
Unlike random integration which often leads to genetic scrambling and necessitates extensive screening to identify functional clones, semi-targeted integration enhances both integration efficiency and expression levels. The transposon as a mobile DNA element, can integrate the entire construct at multiple locations in the genome through a cut-and-paste mechanism. This means that most cells receive a fully functional construct, resulting in higher copy numbers and improved stability of expression. As there is significant reduction in screening number of clones to get the best producer, consequently, the timeline for developing stable cell lines is significantly shortened, enabling researchers to identify high-yield clones more easily with desired qualities. This in turn, has streamlined the path toward successful biopharmaceutical production.