With the use of Liquid Chromatography-Mass Spectrometry (LC-MS) and Gas Chromatography-Mass Spectrometry (GC-MS), impurities are identified very early in the development for process optimization. Relative Response Factor (RRF) is essential to obtain accurate quantification of impurities, when using High Performance Liquid Chromatography (HPLC) method.
The determination of the RRF as it is carried out classically, requires a sample of the isolated impurity. Impurities are usually either synthesized or isolated by preparative HPLC. As this is resource and time-consuming, it is usually carried out late in the development process. However, it is possible to determine RRF of non-isolated impurities by using a combination of HPLC and Nuclear Magnetic Resonance (NMR).
The concept:
Consider a compound and its impurity(imp). The structure and the molecular weight of the impurity have been previously determined by LC-MS.
RRF = Response[compound] = UV area[compound] X conc[imp] (1)
Response[imp] conc[compound] UV area[imp]
Equation 1 shows the classic RRF determination. However, when an impurity and the main compound are present in the same sample, the term concentration can be replaced by weight. The term weight can then be replaced by molecular weight (MW) multiplied by the number of moles(n)
RRF = UV area[compound] x Wt[imp]
Wt[compound] UV area[imp]
Or,
RRF = UV area[compound] x MW[imp] x n[imp] (2)
UV area[imp] MW[compound] n[compound]
In a quantitative 1H NMR spectrum, the resonance integrals, corrected for the number of 1H giving rise to the signal, is directly proportional to the molar concentration. UV peak areas can be obtained from an HPLC chromatogram of a sample containing the main compound and its impurity. Assuming that the molecular weights are known, the RRF can therefore be calculated.
Case Study:
3-cyanobenzaldehyde(1) is one of the four starting materials used in the synthesis of Allyl Hantzsch(2)
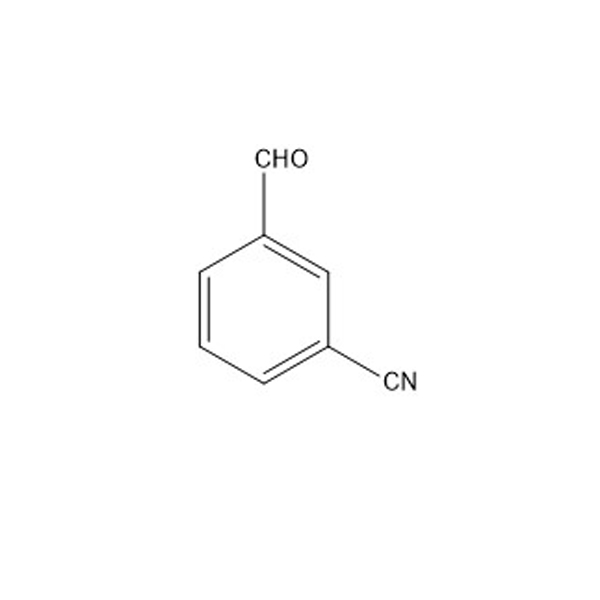
(1)
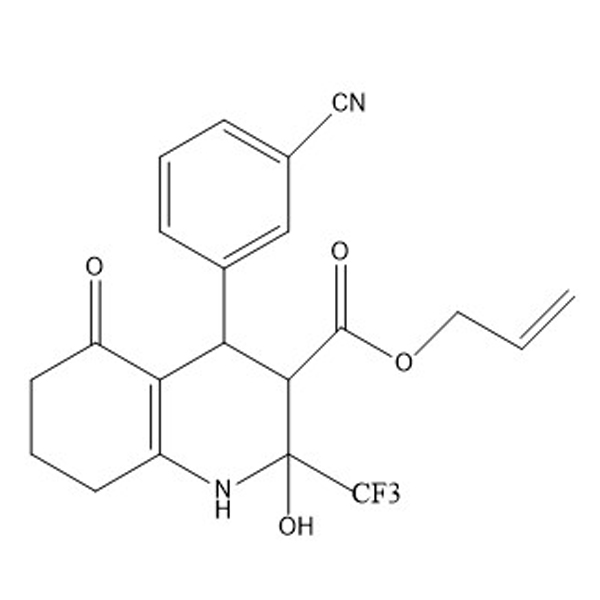
(2)
A sample of Allyl Hantzsch containing 10%w/w of 3-cyanobenzaldehyde was prepared. The sample was diluted in d6-DMSO. An aliquot of the above sample was further diluted with HPLC eluent and a chromatogram was obtained (refer Fig 1). A second aliquot was taken and further diluted with d6-DMSO, and a quantitative 1H NMR spectrum was obtained (refer Fig 2). This was performed in duplicate.
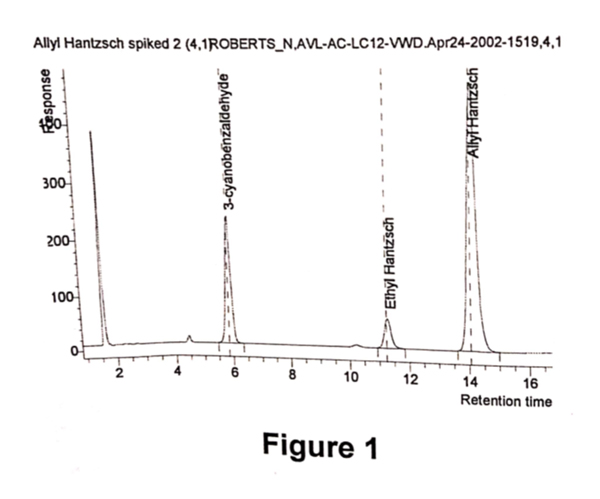
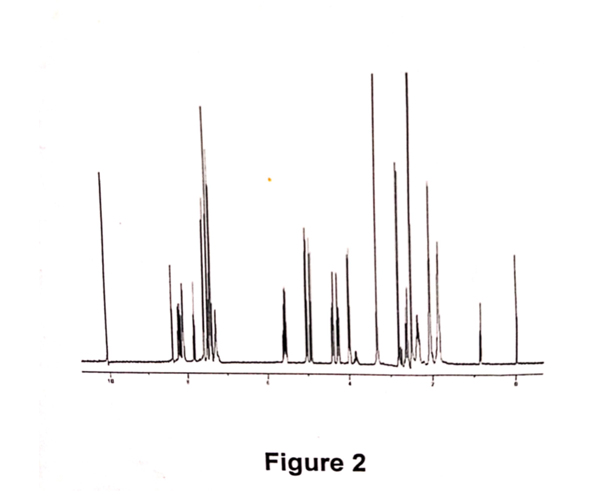
The UV area of Allyl Hantzsch and 3- cyanobenzaldehyde were obtained from the chromatogram, above. The molecular weights are known. The molar ratio was determined by measuring the integral of resonances at 10ppm (aldehyde proton from 3-cyanobenzaldehyde) and at 5pp (allyl proton). Note that, compound and impurity resonances need to be resolved and assigned, and of sufficient signal- to- noise to enable this methodology. Using equation 2, the relative response factor was calculated. As the sample was prepared by weighing a known amount of Allyl Hantzsch and 3-cyanobenzaldehyde, the RRF using equation1 was also determined.
Using the classical determination, the RRF was calculated to be 0.41 and 0.42 for replicate analysis. Using equation2, the RRF was calculated to be 0.37 and 0.37.
Conclusion:
The above case study shows that, using a combination of NMR and HPLC, the RRF of an impurity spike in a sample can be calculated. The RRF obtained by this method is in good agreement with the classically determined value. The determination of RRF of non-isolated impurities using this technique has also been demonstrated.
This approach is a quick and reliable method of determining the RRF of impurities.